| Record Information |
|---|
| Version | 5.0 |
|---|
| Status | Detected and Quantified |
|---|
| Creation Date | 2005-11-16 15:48:42 UTC |
|---|
| Update Date | 2022-03-07 02:48:59 UTC |
|---|
| HMDB ID | HMDB0000036 |
|---|
| Secondary Accession Numbers | |
|---|
| Metabolite Identification |
|---|
| Common Name | Taurocholic acid |
|---|
| Description | Taurocholic acid is a bile acid and is the product of the conjugation of cholic acid with taurine. Its sodium salt is the chief ingredient of the bile of carnivorous animals. Bile acids are steroid acids found predominantly in the bile of mammals. The distinction between different bile acids is minute, depending only on the presence or absence of hydroxyl groups on positions 3, 7, and 12. Bile acids are physiological detergents that facilitate excretion, absorption, and transport of fats and sterols in the intestine and liver. Bile acids are also steroidal amphipathic molecules derived from the catabolism of cholesterol. They modulate bile flow and lipid secretion, are essential for the absorption of dietary fats and vitamins, and have been implicated in the regulation of all the key enzymes involved in cholesterol homeostasis. Bile acids recirculate through the liver, bile ducts, small intestine, and portal vein to form an enterohepatic circuit. They exist as anions at physiological pH, and consequently require a carrier for transport across the membranes of the enterohepatic tissues. The unique detergent properties of bile acids are essential for the digestion and intestinal absorption of hydrophobic nutrients. Bile acids have potent toxic properties (e.g. membrane disruption) and there are a plethora of mechanisms to limit their accumulation in blood and tissues (PMID: 11316487 , 16037564 , 12576301 , 11907135 ). Taurocholic acid, as with all bile acids, acts as a detergent to solubilize fats for absorption and is itself absorbed. It is used as a cholagogue and choleretic (a bile purging agent). Hydrolysis of taurocholic acid yields taurine, a nonessential amino acid. Taurocholic acid is one of the main components of urinary nonsulfated bile acids in biliary atresia. Raised levels of taurocholate in fetal serum in obstetric cholestasis may result in the development of a fetal dysrhythmia and sudden intra-uterine death (PMID: 3944741 , 11256973 ). |
|---|
| Structure | [H][C@@]1(CC[C@@]2([H])[C@]3([H])[C@H](O)C[C@]4([H])C[C@H](O)CC[C@]4(C)[C@@]3([H])C[C@H](O)[C@]12C)[C@H](C)CCC(=O)NCCS(O)(=O)=O InChI=1S/C26H45NO7S/c1-15(4-7-23(31)27-10-11-35(32,33)34)18-5-6-19-24-20(14-22(30)26(18,19)3)25(2)9-8-17(28)12-16(25)13-21(24)29/h15-22,24,28-30H,4-14H2,1-3H3,(H,27,31)(H,32,33,34)/t15-,16+,17-,18-,19+,20+,21-,22+,24+,25+,26-/m1/s1 |
|---|
| Synonyms | | Value | Source |
|---|
| 3alpha,7alpha,12alpha-Trihydroxy-5beta-cholanic acid 24-taurine | ChEBI | | Cholic acid taurine conjugate | ChEBI | | Choloyl-taurine | ChEBI | | Cholyltaurine | ChEBI | | N-Choloyltaurine | ChEBI | | Taurocholate | ChEBI | | 3a,7a,12a-Trihydroxy-5b-cholanate 24-taurine | Generator | | 3a,7a,12a-Trihydroxy-5b-cholanic acid 24-taurine | Generator | | 3alpha,7alpha,12alpha-Trihydroxy-5beta-cholanate 24-taurine | Generator | | 3Α,7α,12α-trihydroxy-5β-cholanate 24-taurine | Generator | | 3Α,7α,12α-trihydroxy-5β-cholanic acid 24-taurine | Generator | | Cholate taurine conjugate | Generator | | Cholic acid taurine conjugic acid | Generator | | Cholaic acid | HMDB | | Cholaate | HMDB | | Taurine cholate | HMDB | | Taurocholic acid, (7 beta)-isomer | HMDB | | Taurocholic acid, (5 alpha)-isomer | HMDB |
|
|---|
| Chemical Formula | C26H45NO7S |
|---|
| Average Molecular Weight | 515.703 |
|---|
| Monoisotopic Molecular Weight | 515.291673489 |
|---|
| IUPAC Name | 2-[(4R)-4-[(1S,2S,5R,7S,9R,10R,11S,14R,15R,16S)-5,9,16-trihydroxy-2,15-dimethyltetracyclo[8.7.0.0^{2,7}.0^{11,15}]heptadecan-14-yl]pentanamido]ethane-1-sulfonic acid |
|---|
| Traditional Name | 2-[(4R)-4-[(1S,2S,5R,7S,9R,10R,11S,14R,15R,16S)-5,9,16-trihydroxy-2,15-dimethyltetracyclo[8.7.0.0^{2,7}.0^{11,15}]heptadecan-14-yl]pentanamido]ethanesulfonic acid |
|---|
| CAS Registry Number | 81-24-3 |
|---|
| SMILES | [H][C@@]1(CC[C@@]2([H])[C@]3([H])[C@H](O)C[C@]4([H])C[C@H](O)CC[C@]4(C)[C@@]3([H])C[C@H](O)[C@]12C)[C@H](C)CCC(=O)NCCS(O)(=O)=O |
|---|
| InChI Identifier | InChI=1S/C26H45NO7S/c1-15(4-7-23(31)27-10-11-35(32,33)34)18-5-6-19-24-20(14-22(30)26(18,19)3)25(2)9-8-17(28)12-16(25)13-21(24)29/h15-22,24,28-30H,4-14H2,1-3H3,(H,27,31)(H,32,33,34)/t15-,16+,17-,18-,19+,20+,21-,22+,24+,25+,26-/m1/s1 |
|---|
| InChI Key | WBWWGRHZICKQGZ-HZAMXZRMSA-N |
|---|
| Chemical Taxonomy |
|---|
| Description | Belongs to the class of organic compounds known as trihydroxy bile acids, alcohols and derivatives. These are prenol lipids structurally characterized by a bile acid or alcohol which bears three hydroxyl groups. |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Lipids and lipid-like molecules |
|---|
| Class | Steroids and steroid derivatives |
|---|
| Sub Class | Bile acids, alcohols and derivatives |
|---|
| Direct Parent | Trihydroxy bile acids, alcohols and derivatives |
|---|
| Alternative Parents | |
|---|
| Substituents | - Trihydroxy bile acid, alcohol, or derivatives
- 3-hydroxysteroid
- 12-hydroxysteroid
- Hydroxysteroid
- 3-alpha-hydroxysteroid
- 7-hydroxysteroid
- Cyclic alcohol
- Organic sulfonic acid or derivatives
- Organosulfonic acid or derivatives
- Organosulfonic acid
- Sulfonyl
- Alkanesulfonic acid
- Secondary alcohol
- Carboximidic acid
- Polyol
- Carboximidic acid derivative
- Organic 1,3-dipolar compound
- Propargyl-type 1,3-dipolar organic compound
- Alcohol
- Organosulfur compound
- Organooxygen compound
- Organonitrogen compound
- Organopnictogen compound
- Organic oxygen compound
- Organic oxide
- Hydrocarbon derivative
- Organic nitrogen compound
- Aliphatic homopolycyclic compound
|
|---|
| Molecular Framework | Aliphatic homopolycyclic compounds |
|---|
| External Descriptors | |
|---|
| Ontology |
|---|
| Not Available | Not Available |
|---|
| Physical Properties |
|---|
| State | Solid |
|---|
| Experimental Molecular Properties | | Property | Value | Reference |
|---|
| Melting Point | 125 °C | Not Available | | Boiling Point | Not Available | Not Available | | Water Solubility | Not Available | Not Available | | LogP | Not Available | Not Available |
|
|---|
| Experimental Chromatographic Properties | Not Available |
|---|
| Predicted Molecular Properties | |
|---|
| Predicted Chromatographic Properties | Predicted Collision Cross SectionsPredicted Kovats Retention IndicesUnderivatizedDerivatized| Derivative Name / Structure | SMILES | Kovats RI Value | Column Type | Reference |
|---|
| Taurocholic acid,1TMS,isomer #1 | C[C@H](CCC(=O)NCCS(=O)(=O)O)[C@H]1CC[C@H]2[C@H]3[C@H](C[C@H](O)[C@@]21C)[C@@]1(C)CC[C@@H](O)C[C@H]1C[C@H]3O[Si](C)(C)C | 4179.1 | Semi standard non polar | 33892256 | | Taurocholic acid,1TMS,isomer #2 | C[C@H](CCC(=O)NCCS(=O)(=O)O)[C@H]1CC[C@H]2[C@H]3[C@H](C[C@H](O)[C@@]21C)[C@@]1(C)CC[C@@H](O[Si](C)(C)C)C[C@H]1C[C@H]3O | 4303.3 | Semi standard non polar | 33892256 | | Taurocholic acid,1TMS,isomer #3 | C[C@H](CCC(=O)NCCS(=O)(=O)O)[C@H]1CC[C@H]2[C@H]3[C@H](C[C@H](O[Si](C)(C)C)[C@@]21C)[C@@]1(C)CC[C@@H](O)C[C@H]1C[C@H]3O | 4263.1 | Semi standard non polar | 33892256 | | Taurocholic acid,1TMS,isomer #4 | C[C@H](CCC(=O)NCCS(=O)(=O)O[Si](C)(C)C)[C@H]1CC[C@H]2[C@H]3[C@H](C[C@H](O)[C@@]21C)[C@@]1(C)CC[C@@H](O)C[C@H]1C[C@H]3O | 4404.5 | Semi standard non polar | 33892256 | | Taurocholic acid,1TMS,isomer #5 | C[C@H](CCC(=O)N(CCS(=O)(=O)O)[Si](C)(C)C)[C@H]1CC[C@H]2[C@H]3[C@H](C[C@H](O)[C@@]21C)[C@@]1(C)CC[C@@H](O)C[C@H]1C[C@H]3O | 4295.1 | Semi standard non polar | 33892256 | | Taurocholic acid,2TMS,isomer #1 | C[C@H](CCC(=O)NCCS(=O)(=O)O)[C@H]1CC[C@H]2[C@H]3[C@H](C[C@H](O[Si](C)(C)C)[C@@]21C)[C@@]1(C)CC[C@@H](O)C[C@H]1C[C@H]3O[Si](C)(C)C | 4119.4 | Semi standard non polar | 33892256 | | Taurocholic acid,2TMS,isomer #10 | C[C@H](CCC(=O)N(CCS(=O)(=O)O[Si](C)(C)C)[Si](C)(C)C)[C@H]1CC[C@H]2[C@H]3[C@H](C[C@H](O)[C@@]21C)[C@@]1(C)CC[C@@H](O)C[C@H]1C[C@H]3O | 4379.6 | Semi standard non polar | 33892256 | | Taurocholic acid,2TMS,isomer #2 | C[C@H](CCC(=O)NCCS(=O)(=O)O)[C@H]1CC[C@H]2[C@H]3[C@H](C[C@H](O)[C@@]21C)[C@@]1(C)CC[C@@H](O[Si](C)(C)C)C[C@H]1C[C@H]3O[Si](C)(C)C | 4141.6 | Semi standard non polar | 33892256 | | Taurocholic acid,2TMS,isomer #3 | C[C@H](CCC(=O)NCCS(=O)(=O)O[Si](C)(C)C)[C@H]1CC[C@H]2[C@H]3[C@H](C[C@H](O)[C@@]21C)[C@@]1(C)CC[C@@H](O)C[C@H]1C[C@H]3O[Si](C)(C)C | 4216.6 | Semi standard non polar | 33892256 | | Taurocholic acid,2TMS,isomer #4 | C[C@H](CCC(=O)N(CCS(=O)(=O)O)[Si](C)(C)C)[C@H]1CC[C@H]2[C@H]3[C@H](C[C@H](O)[C@@]21C)[C@@]1(C)CC[C@@H](O)C[C@H]1C[C@H]3O[Si](C)(C)C | 4103.9 | Semi standard non polar | 33892256 | | Taurocholic acid,2TMS,isomer #5 | C[C@H](CCC(=O)NCCS(=O)(=O)O)[C@H]1CC[C@H]2[C@H]3[C@H](C[C@H](O[Si](C)(C)C)[C@@]21C)[C@@]1(C)CC[C@@H](O[Si](C)(C)C)C[C@H]1C[C@H]3O | 4181.6 | Semi standard non polar | 33892256 | | Taurocholic acid,2TMS,isomer #6 | C[C@H](CCC(=O)NCCS(=O)(=O)O[Si](C)(C)C)[C@H]1CC[C@H]2[C@H]3[C@H](C[C@H](O)[C@@]21C)[C@@]1(C)CC[C@@H](O[Si](C)(C)C)C[C@H]1C[C@H]3O | 4359.2 | Semi standard non polar | 33892256 | | Taurocholic acid,2TMS,isomer #7 | C[C@H](CCC(=O)N(CCS(=O)(=O)O)[Si](C)(C)C)[C@H]1CC[C@H]2[C@H]3[C@H](C[C@H](O)[C@@]21C)[C@@]1(C)CC[C@@H](O[Si](C)(C)C)C[C@H]1C[C@H]3O | 4248.7 | Semi standard non polar | 33892256 | | Taurocholic acid,2TMS,isomer #8 | C[C@H](CCC(=O)NCCS(=O)(=O)O[Si](C)(C)C)[C@H]1CC[C@H]2[C@H]3[C@H](C[C@H](O[Si](C)(C)C)[C@@]21C)[C@@]1(C)CC[C@@H](O)C[C@H]1C[C@H]3O | 4304.0 | Semi standard non polar | 33892256 | | Taurocholic acid,2TMS,isomer #9 | C[C@H](CCC(=O)N(CCS(=O)(=O)O)[Si](C)(C)C)[C@H]1CC[C@H]2[C@H]3[C@H](C[C@H](O[Si](C)(C)C)[C@@]21C)[C@@]1(C)CC[C@@H](O)C[C@H]1C[C@H]3O | 4184.3 | Semi standard non polar | 33892256 | | Taurocholic acid,3TMS,isomer #1 | C[C@H](CCC(=O)NCCS(=O)(=O)O)[C@H]1CC[C@H]2[C@H]3[C@H](C[C@H](O[Si](C)(C)C)[C@@]21C)[C@@]1(C)CC[C@@H](O[Si](C)(C)C)C[C@H]1C[C@H]3O[Si](C)(C)C | 4087.3 | Semi standard non polar | 33892256 | | Taurocholic acid,3TMS,isomer #10 | C[C@H](CCC(=O)N(CCS(=O)(=O)O[Si](C)(C)C)[Si](C)(C)C)[C@H]1CC[C@H]2[C@H]3[C@H](C[C@H](O[Si](C)(C)C)[C@@]21C)[C@@]1(C)CC[C@@H](O)C[C@H]1C[C@H]3O | 4239.7 | Semi standard non polar | 33892256 | | Taurocholic acid,3TMS,isomer #2 | C[C@H](CCC(=O)NCCS(=O)(=O)O[Si](C)(C)C)[C@H]1CC[C@H]2[C@H]3[C@H](C[C@H](O[Si](C)(C)C)[C@@]21C)[C@@]1(C)CC[C@@H](O)C[C@H]1C[C@H]3O[Si](C)(C)C | 4164.5 | Semi standard non polar | 33892256 | | Taurocholic acid,3TMS,isomer #3 | C[C@H](CCC(=O)N(CCS(=O)(=O)O)[Si](C)(C)C)[C@H]1CC[C@H]2[C@H]3[C@H](C[C@H](O[Si](C)(C)C)[C@@]21C)[C@@]1(C)CC[C@@H](O)C[C@H]1C[C@H]3O[Si](C)(C)C | 4054.4 | Semi standard non polar | 33892256 | | Taurocholic acid,3TMS,isomer #4 | C[C@H](CCC(=O)NCCS(=O)(=O)O[Si](C)(C)C)[C@H]1CC[C@H]2[C@H]3[C@H](C[C@H](O)[C@@]21C)[C@@]1(C)CC[C@@H](O[Si](C)(C)C)C[C@H]1C[C@H]3O[Si](C)(C)C | 4180.1 | Semi standard non polar | 33892256 | | Taurocholic acid,3TMS,isomer #5 | C[C@H](CCC(=O)N(CCS(=O)(=O)O)[Si](C)(C)C)[C@H]1CC[C@H]2[C@H]3[C@H](C[C@H](O)[C@@]21C)[C@@]1(C)CC[C@@H](O[Si](C)(C)C)C[C@H]1C[C@H]3O[Si](C)(C)C | 4069.1 | Semi standard non polar | 33892256 | | Taurocholic acid,3TMS,isomer #6 | C[C@H](CCC(=O)N(CCS(=O)(=O)O[Si](C)(C)C)[Si](C)(C)C)[C@H]1CC[C@H]2[C@H]3[C@H](C[C@H](O)[C@@]21C)[C@@]1(C)CC[C@@H](O)C[C@H]1C[C@H]3O[Si](C)(C)C | 4170.4 | Semi standard non polar | 33892256 | | Taurocholic acid,3TMS,isomer #7 | C[C@H](CCC(=O)NCCS(=O)(=O)O[Si](C)(C)C)[C@H]1CC[C@H]2[C@H]3[C@H](C[C@H](O[Si](C)(C)C)[C@@]21C)[C@@]1(C)CC[C@@H](O[Si](C)(C)C)C[C@H]1C[C@H]3O | 4209.1 | Semi standard non polar | 33892256 | | Taurocholic acid,3TMS,isomer #8 | C[C@H](CCC(=O)N(CCS(=O)(=O)O)[Si](C)(C)C)[C@H]1CC[C@H]2[C@H]3[C@H](C[C@H](O[Si](C)(C)C)[C@@]21C)[C@@]1(C)CC[C@@H](O[Si](C)(C)C)C[C@H]1C[C@H]3O | 4106.4 | Semi standard non polar | 33892256 | | Taurocholic acid,3TMS,isomer #9 | C[C@H](CCC(=O)N(CCS(=O)(=O)O[Si](C)(C)C)[Si](C)(C)C)[C@H]1CC[C@H]2[C@H]3[C@H](C[C@H](O)[C@@]21C)[C@@]1(C)CC[C@@H](O[Si](C)(C)C)C[C@H]1C[C@H]3O | 4299.9 | Semi standard non polar | 33892256 | | Taurocholic acid,4TMS,isomer #1 | C[C@H](CCC(=O)NCCS(=O)(=O)O[Si](C)(C)C)[C@H]1CC[C@H]2[C@H]3[C@H](C[C@H](O[Si](C)(C)C)[C@@]21C)[C@@]1(C)CC[C@@H](O[Si](C)(C)C)C[C@H]1C[C@H]3O[Si](C)(C)C | 4118.0 | Semi standard non polar | 33892256 | | Taurocholic acid,4TMS,isomer #1 | C[C@H](CCC(=O)NCCS(=O)(=O)O[Si](C)(C)C)[C@H]1CC[C@H]2[C@H]3[C@H](C[C@H](O[Si](C)(C)C)[C@@]21C)[C@@]1(C)CC[C@@H](O[Si](C)(C)C)C[C@H]1C[C@H]3O[Si](C)(C)C | 4654.9 | Standard non polar | 33892256 | | Taurocholic acid,4TMS,isomer #1 | C[C@H](CCC(=O)NCCS(=O)(=O)O[Si](C)(C)C)[C@H]1CC[C@H]2[C@H]3[C@H](C[C@H](O[Si](C)(C)C)[C@@]21C)[C@@]1(C)CC[C@@H](O[Si](C)(C)C)C[C@H]1C[C@H]3O[Si](C)(C)C | 4734.5 | Standard polar | 33892256 | | Taurocholic acid,4TMS,isomer #2 | C[C@H](CCC(=O)N(CCS(=O)(=O)O)[Si](C)(C)C)[C@H]1CC[C@H]2[C@H]3[C@H](C[C@H](O[Si](C)(C)C)[C@@]21C)[C@@]1(C)CC[C@@H](O[Si](C)(C)C)C[C@H]1C[C@H]3O[Si](C)(C)C | 4033.4 | Semi standard non polar | 33892256 | | Taurocholic acid,4TMS,isomer #2 | C[C@H](CCC(=O)N(CCS(=O)(=O)O)[Si](C)(C)C)[C@H]1CC[C@H]2[C@H]3[C@H](C[C@H](O[Si](C)(C)C)[C@@]21C)[C@@]1(C)CC[C@@H](O[Si](C)(C)C)C[C@H]1C[C@H]3O[Si](C)(C)C | 4666.6 | Standard non polar | 33892256 | | Taurocholic acid,4TMS,isomer #2 | C[C@H](CCC(=O)N(CCS(=O)(=O)O)[Si](C)(C)C)[C@H]1CC[C@H]2[C@H]3[C@H](C[C@H](O[Si](C)(C)C)[C@@]21C)[C@@]1(C)CC[C@@H](O[Si](C)(C)C)C[C@H]1C[C@H]3O[Si](C)(C)C | 4770.6 | Standard polar | 33892256 | | Taurocholic acid,4TMS,isomer #3 | C[C@H](CCC(=O)N(CCS(=O)(=O)O[Si](C)(C)C)[Si](C)(C)C)[C@H]1CC[C@H]2[C@H]3[C@H](C[C@H](O[Si](C)(C)C)[C@@]21C)[C@@]1(C)CC[C@@H](O)C[C@H]1C[C@H]3O[Si](C)(C)C | 4132.0 | Semi standard non polar | 33892256 | | Taurocholic acid,4TMS,isomer #3 | C[C@H](CCC(=O)N(CCS(=O)(=O)O[Si](C)(C)C)[Si](C)(C)C)[C@H]1CC[C@H]2[C@H]3[C@H](C[C@H](O[Si](C)(C)C)[C@@]21C)[C@@]1(C)CC[C@@H](O)C[C@H]1C[C@H]3O[Si](C)(C)C | 4752.4 | Standard non polar | 33892256 | | Taurocholic acid,4TMS,isomer #3 | C[C@H](CCC(=O)N(CCS(=O)(=O)O[Si](C)(C)C)[Si](C)(C)C)[C@H]1CC[C@H]2[C@H]3[C@H](C[C@H](O[Si](C)(C)C)[C@@]21C)[C@@]1(C)CC[C@@H](O)C[C@H]1C[C@H]3O[Si](C)(C)C | 4721.7 | Standard polar | 33892256 | | Taurocholic acid,4TMS,isomer #4 | C[C@H](CCC(=O)N(CCS(=O)(=O)O[Si](C)(C)C)[Si](C)(C)C)[C@H]1CC[C@H]2[C@H]3[C@H](C[C@H](O)[C@@]21C)[C@@]1(C)CC[C@@H](O[Si](C)(C)C)C[C@H]1C[C@H]3O[Si](C)(C)C | 4146.3 | Semi standard non polar | 33892256 | | Taurocholic acid,4TMS,isomer #4 | C[C@H](CCC(=O)N(CCS(=O)(=O)O[Si](C)(C)C)[Si](C)(C)C)[C@H]1CC[C@H]2[C@H]3[C@H](C[C@H](O)[C@@]21C)[C@@]1(C)CC[C@@H](O[Si](C)(C)C)C[C@H]1C[C@H]3O[Si](C)(C)C | 4742.5 | Standard non polar | 33892256 | | Taurocholic acid,4TMS,isomer #4 | C[C@H](CCC(=O)N(CCS(=O)(=O)O[Si](C)(C)C)[Si](C)(C)C)[C@H]1CC[C@H]2[C@H]3[C@H](C[C@H](O)[C@@]21C)[C@@]1(C)CC[C@@H](O[Si](C)(C)C)C[C@H]1C[C@H]3O[Si](C)(C)C | 4774.7 | Standard polar | 33892256 | | Taurocholic acid,4TMS,isomer #5 | C[C@H](CCC(=O)N(CCS(=O)(=O)O[Si](C)(C)C)[Si](C)(C)C)[C@H]1CC[C@H]2[C@H]3[C@H](C[C@H](O[Si](C)(C)C)[C@@]21C)[C@@]1(C)CC[C@@H](O[Si](C)(C)C)C[C@H]1C[C@H]3O | 4165.2 | Semi standard non polar | 33892256 | | Taurocholic acid,4TMS,isomer #5 | C[C@H](CCC(=O)N(CCS(=O)(=O)O[Si](C)(C)C)[Si](C)(C)C)[C@H]1CC[C@H]2[C@H]3[C@H](C[C@H](O[Si](C)(C)C)[C@@]21C)[C@@]1(C)CC[C@@H](O[Si](C)(C)C)C[C@H]1C[C@H]3O | 4736.0 | Standard non polar | 33892256 | | Taurocholic acid,4TMS,isomer #5 | C[C@H](CCC(=O)N(CCS(=O)(=O)O[Si](C)(C)C)[Si](C)(C)C)[C@H]1CC[C@H]2[C@H]3[C@H](C[C@H](O[Si](C)(C)C)[C@@]21C)[C@@]1(C)CC[C@@H](O[Si](C)(C)C)C[C@H]1C[C@H]3O | 4697.5 | Standard polar | 33892256 | | Taurocholic acid,5TMS,isomer #1 | C[C@H](CCC(=O)N(CCS(=O)(=O)O[Si](C)(C)C)[Si](C)(C)C)[C@H]1CC[C@H]2[C@H]3[C@H](C[C@H](O[Si](C)(C)C)[C@@]21C)[C@@]1(C)CC[C@@H](O[Si](C)(C)C)C[C@H]1C[C@H]3O[Si](C)(C)C | 4100.9 | Semi standard non polar | 33892256 | | Taurocholic acid,5TMS,isomer #1 | C[C@H](CCC(=O)N(CCS(=O)(=O)O[Si](C)(C)C)[Si](C)(C)C)[C@H]1CC[C@H]2[C@H]3[C@H](C[C@H](O[Si](C)(C)C)[C@@]21C)[C@@]1(C)CC[C@@H](O[Si](C)(C)C)C[C@H]1C[C@H]3O[Si](C)(C)C | 4861.3 | Standard non polar | 33892256 | | Taurocholic acid,5TMS,isomer #1 | C[C@H](CCC(=O)N(CCS(=O)(=O)O[Si](C)(C)C)[Si](C)(C)C)[C@H]1CC[C@H]2[C@H]3[C@H](C[C@H](O[Si](C)(C)C)[C@@]21C)[C@@]1(C)CC[C@@H](O[Si](C)(C)C)C[C@H]1C[C@H]3O[Si](C)(C)C | 4555.7 | Standard polar | 33892256 | | Taurocholic acid,1TBDMS,isomer #1 | C[C@H](CCC(=O)NCCS(=O)(=O)O)[C@H]1CC[C@H]2[C@H]3[C@H](C[C@H](O)[C@@]21C)[C@@]1(C)CC[C@@H](O)C[C@H]1C[C@H]3O[Si](C)(C)C(C)(C)C | 4386.3 | Semi standard non polar | 33892256 | | Taurocholic acid,1TBDMS,isomer #2 | C[C@H](CCC(=O)NCCS(=O)(=O)O)[C@H]1CC[C@H]2[C@H]3[C@H](C[C@H](O)[C@@]21C)[C@@]1(C)CC[C@@H](O[Si](C)(C)C(C)(C)C)C[C@H]1C[C@H]3O | 4518.0 | Semi standard non polar | 33892256 | | Taurocholic acid,1TBDMS,isomer #3 | C[C@H](CCC(=O)NCCS(=O)(=O)O)[C@H]1CC[C@H]2[C@H]3[C@H](C[C@H](O[Si](C)(C)C(C)(C)C)[C@@]21C)[C@@]1(C)CC[C@@H](O)C[C@H]1C[C@H]3O | 4463.2 | Semi standard non polar | 33892256 | | Taurocholic acid,1TBDMS,isomer #4 | C[C@H](CCC(=O)NCCS(=O)(=O)O[Si](C)(C)C(C)(C)C)[C@H]1CC[C@H]2[C@H]3[C@H](C[C@H](O)[C@@]21C)[C@@]1(C)CC[C@@H](O)C[C@H]1C[C@H]3O | 4607.1 | Semi standard non polar | 33892256 | | Taurocholic acid,1TBDMS,isomer #5 | C[C@H](CCC(=O)N(CCS(=O)(=O)O)[Si](C)(C)C(C)(C)C)[C@H]1CC[C@H]2[C@H]3[C@H](C[C@H](O)[C@@]21C)[C@@]1(C)CC[C@@H](O)C[C@H]1C[C@H]3O | 4569.9 | Semi standard non polar | 33892256 | | Taurocholic acid,2TBDMS,isomer #1 | C[C@H](CCC(=O)NCCS(=O)(=O)O)[C@H]1CC[C@H]2[C@H]3[C@H](C[C@H](O[Si](C)(C)C(C)(C)C)[C@@]21C)[C@@]1(C)CC[C@@H](O)C[C@H]1C[C@H]3O[Si](C)(C)C(C)(C)C | 4539.7 | Semi standard non polar | 33892256 | | Taurocholic acid,2TBDMS,isomer #10 | C[C@H](CCC(=O)N(CCS(=O)(=O)O[Si](C)(C)C(C)(C)C)[Si](C)(C)C(C)(C)C)[C@H]1CC[C@H]2[C@H]3[C@H](C[C@H](O)[C@@]21C)[C@@]1(C)CC[C@@H](O)C[C@H]1C[C@H]3O | 4848.1 | Semi standard non polar | 33892256 | | Taurocholic acid,2TBDMS,isomer #2 | C[C@H](CCC(=O)NCCS(=O)(=O)O)[C@H]1CC[C@H]2[C@H]3[C@H](C[C@H](O)[C@@]21C)[C@@]1(C)CC[C@@H](O[Si](C)(C)C(C)(C)C)C[C@H]1C[C@H]3O[Si](C)(C)C(C)(C)C | 4559.2 | Semi standard non polar | 33892256 | | Taurocholic acid,2TBDMS,isomer #3 | C[C@H](CCC(=O)NCCS(=O)(=O)O[Si](C)(C)C(C)(C)C)[C@H]1CC[C@H]2[C@H]3[C@H](C[C@H](O)[C@@]21C)[C@@]1(C)CC[C@@H](O)C[C@H]1C[C@H]3O[Si](C)(C)C(C)(C)C | 4632.6 | Semi standard non polar | 33892256 | | Taurocholic acid,2TBDMS,isomer #4 | C[C@H](CCC(=O)N(CCS(=O)(=O)O)[Si](C)(C)C(C)(C)C)[C@H]1CC[C@H]2[C@H]3[C@H](C[C@H](O)[C@@]21C)[C@@]1(C)CC[C@@H](O)C[C@H]1C[C@H]3O[Si](C)(C)C(C)(C)C | 4589.8 | Semi standard non polar | 33892256 | | Taurocholic acid,2TBDMS,isomer #5 | C[C@H](CCC(=O)NCCS(=O)(=O)O)[C@H]1CC[C@H]2[C@H]3[C@H](C[C@H](O[Si](C)(C)C(C)(C)C)[C@@]21C)[C@@]1(C)CC[C@@H](O[Si](C)(C)C(C)(C)C)C[C@H]1C[C@H]3O | 4595.2 | Semi standard non polar | 33892256 | | Taurocholic acid,2TBDMS,isomer #6 | C[C@H](CCC(=O)NCCS(=O)(=O)O[Si](C)(C)C(C)(C)C)[C@H]1CC[C@H]2[C@H]3[C@H](C[C@H](O)[C@@]21C)[C@@]1(C)CC[C@@H](O[Si](C)(C)C(C)(C)C)C[C@H]1C[C@H]3O | 4800.5 | Semi standard non polar | 33892256 | | Taurocholic acid,2TBDMS,isomer #7 | C[C@H](CCC(=O)N(CCS(=O)(=O)O)[Si](C)(C)C(C)(C)C)[C@H]1CC[C@H]2[C@H]3[C@H](C[C@H](O)[C@@]21C)[C@@]1(C)CC[C@@H](O[Si](C)(C)C(C)(C)C)C[C@H]1C[C@H]3O | 4727.0 | Semi standard non polar | 33892256 | | Taurocholic acid,2TBDMS,isomer #8 | C[C@H](CCC(=O)NCCS(=O)(=O)O[Si](C)(C)C(C)(C)C)[C@H]1CC[C@H]2[C@H]3[C@H](C[C@H](O[Si](C)(C)C(C)(C)C)[C@@]21C)[C@@]1(C)CC[C@@H](O)C[C@H]1C[C@H]3O | 4723.3 | Semi standard non polar | 33892256 | | Taurocholic acid,2TBDMS,isomer #9 | C[C@H](CCC(=O)N(CCS(=O)(=O)O)[Si](C)(C)C(C)(C)C)[C@H]1CC[C@H]2[C@H]3[C@H](C[C@H](O[Si](C)(C)C(C)(C)C)[C@@]21C)[C@@]1(C)CC[C@@H](O)C[C@H]1C[C@H]3O | 4658.1 | Semi standard non polar | 33892256 | | Taurocholic acid,3TBDMS,isomer #1 | C[C@H](CCC(=O)NCCS(=O)(=O)O)[C@H]1CC[C@H]2[C@H]3[C@H](C[C@H](O[Si](C)(C)C(C)(C)C)[C@@]21C)[C@@]1(C)CC[C@@H](O[Si](C)(C)C(C)(C)C)C[C@H]1C[C@H]3O[Si](C)(C)C(C)(C)C | 4692.2 | Semi standard non polar | 33892256 | | Taurocholic acid,3TBDMS,isomer #10 | C[C@H](CCC(=O)N(CCS(=O)(=O)O[Si](C)(C)C(C)(C)C)[Si](C)(C)C(C)(C)C)[C@H]1CC[C@H]2[C@H]3[C@H](C[C@H](O[Si](C)(C)C(C)(C)C)[C@@]21C)[C@@]1(C)CC[C@@H](O)C[C@H]1C[C@H]3O | 4920.8 | Semi standard non polar | 33892256 | | Taurocholic acid,3TBDMS,isomer #2 | C[C@H](CCC(=O)NCCS(=O)(=O)O[Si](C)(C)C(C)(C)C)[C@H]1CC[C@H]2[C@H]3[C@H](C[C@H](O[Si](C)(C)C(C)(C)C)[C@@]21C)[C@@]1(C)CC[C@@H](O)C[C@H]1C[C@H]3O[Si](C)(C)C(C)(C)C | 4767.8 | Semi standard non polar | 33892256 | | Taurocholic acid,3TBDMS,isomer #3 | C[C@H](CCC(=O)N(CCS(=O)(=O)O)[Si](C)(C)C(C)(C)C)[C@H]1CC[C@H]2[C@H]3[C@H](C[C@H](O[Si](C)(C)C(C)(C)C)[C@@]21C)[C@@]1(C)CC[C@@H](O)C[C@H]1C[C@H]3O[Si](C)(C)C(C)(C)C | 4729.7 | Semi standard non polar | 33892256 | | Taurocholic acid,3TBDMS,isomer #4 | C[C@H](CCC(=O)NCCS(=O)(=O)O[Si](C)(C)C(C)(C)C)[C@H]1CC[C@H]2[C@H]3[C@H](C[C@H](O)[C@@]21C)[C@@]1(C)CC[C@@H](O[Si](C)(C)C(C)(C)C)C[C@H]1C[C@H]3O[Si](C)(C)C(C)(C)C | 4780.8 | Semi standard non polar | 33892256 | | Taurocholic acid,3TBDMS,isomer #5 | C[C@H](CCC(=O)N(CCS(=O)(=O)O)[Si](C)(C)C(C)(C)C)[C@H]1CC[C@H]2[C@H]3[C@H](C[C@H](O)[C@@]21C)[C@@]1(C)CC[C@@H](O[Si](C)(C)C(C)(C)C)C[C@H]1C[C@H]3O[Si](C)(C)C(C)(C)C | 4752.2 | Semi standard non polar | 33892256 | | Taurocholic acid,3TBDMS,isomer #6 | C[C@H](CCC(=O)N(CCS(=O)(=O)O[Si](C)(C)C(C)(C)C)[Si](C)(C)C(C)(C)C)[C@H]1CC[C@H]2[C@H]3[C@H](C[C@H](O)[C@@]21C)[C@@]1(C)CC[C@@H](O)C[C@H]1C[C@H]3O[Si](C)(C)C(C)(C)C | 4839.5 | Semi standard non polar | 33892256 | | Taurocholic acid,3TBDMS,isomer #7 | C[C@H](CCC(=O)NCCS(=O)(=O)O[Si](C)(C)C(C)(C)C)[C@H]1CC[C@H]2[C@H]3[C@H](C[C@H](O[Si](C)(C)C(C)(C)C)[C@@]21C)[C@@]1(C)CC[C@@H](O[Si](C)(C)C(C)(C)C)C[C@H]1C[C@H]3O | 4813.5 | Semi standard non polar | 33892256 | | Taurocholic acid,3TBDMS,isomer #8 | C[C@H](CCC(=O)N(CCS(=O)(=O)O)[Si](C)(C)C(C)(C)C)[C@H]1CC[C@H]2[C@H]3[C@H](C[C@H](O[Si](C)(C)C(C)(C)C)[C@@]21C)[C@@]1(C)CC[C@@H](O[Si](C)(C)C(C)(C)C)C[C@H]1C[C@H]3O | 4788.6 | Semi standard non polar | 33892256 | | Taurocholic acid,3TBDMS,isomer #9 | C[C@H](CCC(=O)N(CCS(=O)(=O)O[Si](C)(C)C(C)(C)C)[Si](C)(C)C(C)(C)C)[C@H]1CC[C@H]2[C@H]3[C@H](C[C@H](O)[C@@]21C)[C@@]1(C)CC[C@@H](O[Si](C)(C)C(C)(C)C)C[C@H]1C[C@H]3O | 4981.2 | Semi standard non polar | 33892256 |
|
|---|
| Spectra |
|---|
| GC-MS Spectra| Spectrum Type | Description | Splash Key | Deposition Date | Source | View |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - Taurocholic acid GC-MS (TMS_1_1) - 70eV, Positive | Not Available | 2021-11-05 | Wishart Lab | View Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - Taurocholic acid GC-MS (TMS_1_2) - 70eV, Positive | Not Available | 2021-11-05 | Wishart Lab | View Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - Taurocholic acid GC-MS (TMS_1_3) - 70eV, Positive | Not Available | 2021-11-05 | Wishart Lab | View Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - Taurocholic acid GC-MS (TMS_1_4) - 70eV, Positive | Not Available | 2021-11-05 | Wishart Lab | View Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - Taurocholic acid GC-MS (TMS_1_5) - 70eV, Positive | Not Available | 2021-11-05 | Wishart Lab | View Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - Taurocholic acid GC-MS (TMS_2_1) - 70eV, Positive | Not Available | 2021-11-05 | Wishart Lab | View Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - Taurocholic acid GC-MS (TMS_2_2) - 70eV, Positive | Not Available | 2021-11-05 | Wishart Lab | View Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - Taurocholic acid GC-MS (TMS_2_3) - 70eV, Positive | Not Available | 2021-11-05 | Wishart Lab | View Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - Taurocholic acid GC-MS (TMS_2_4) - 70eV, Positive | Not Available | 2021-11-05 | Wishart Lab | View Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - Taurocholic acid GC-MS (TMS_2_5) - 70eV, Positive | Not Available | 2021-11-05 | Wishart Lab | View Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - Taurocholic acid GC-MS (TMS_2_6) - 70eV, Positive | Not Available | 2021-11-05 | Wishart Lab | View Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - Taurocholic acid GC-MS (TMS_2_7) - 70eV, Positive | Not Available | 2021-11-05 | Wishart Lab | View Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - Taurocholic acid GC-MS (TMS_2_8) - 70eV, Positive | Not Available | 2021-11-05 | Wishart Lab | View Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - Taurocholic acid GC-MS (TMS_2_9) - 70eV, Positive | Not Available | 2021-11-05 | Wishart Lab | View Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - Taurocholic acid GC-MS (TMS_2_10) - 70eV, Positive | Not Available | 2021-11-05 | Wishart Lab | View Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - Taurocholic acid GC-MS (TMS_3_1) - 70eV, Positive | Not Available | 2021-11-05 | Wishart Lab | View Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - Taurocholic acid GC-MS (TMS_3_2) - 70eV, Positive | Not Available | 2021-11-05 | Wishart Lab | View Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - Taurocholic acid GC-MS (TMS_3_3) - 70eV, Positive | Not Available | 2021-11-05 | Wishart Lab | View Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - Taurocholic acid GC-MS (TMS_3_4) - 70eV, Positive | Not Available | 2021-11-05 | Wishart Lab | View Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - Taurocholic acid GC-MS (TMS_3_5) - 70eV, Positive | Not Available | 2021-11-05 | Wishart Lab | View Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - Taurocholic acid GC-MS (TMS_3_6) - 70eV, Positive | Not Available | 2021-11-05 | Wishart Lab | View Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - Taurocholic acid GC-MS (TMS_3_7) - 70eV, Positive | Not Available | 2021-11-05 | Wishart Lab | View Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - Taurocholic acid GC-MS (TMS_3_8) - 70eV, Positive | Not Available | 2021-11-05 | Wishart Lab | View Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - Taurocholic acid GC-MS (TMS_3_9) - 70eV, Positive | Not Available | 2021-11-05 | Wishart Lab | View Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - Taurocholic acid GC-MS (TMS_3_10) - 70eV, Positive | Not Available | 2021-11-05 | Wishart Lab | View Spectrum |
MS/MS Spectra| Spectrum Type | Description | Splash Key | Deposition Date | Source | View |
|---|
| Experimental LC-MS/MS | LC-MS/MS Spectrum - Taurocholic acid LC-ESI-QQ , negative-QTOF | splash10-03di-0000090000-911b2f8adf5d8a253a26 | 2017-09-14 | HMDB team, MONA | View Spectrum | | Experimental LC-MS/MS | LC-MS/MS Spectrum - Taurocholic acid LC-ESI-QQ , negative-QTOF | splash10-03di-0000090000-229891181661857b574b | 2017-09-14 | HMDB team, MONA | View Spectrum | | Experimental LC-MS/MS | LC-MS/MS Spectrum - Taurocholic acid LC-ESI-QQ , negative-QTOF | splash10-03di-0000090000-e3b432e2a36b91fb1106 | 2017-09-14 | HMDB team, MONA | View Spectrum | | Experimental LC-MS/MS | LC-MS/MS Spectrum - Taurocholic acid LC-ESI-QQ , negative-QTOF | splash10-03di-0000090000-afa59839c0e5c1b1f875 | 2017-09-14 | HMDB team, MONA | View Spectrum | | Experimental LC-MS/MS | LC-MS/MS Spectrum - Taurocholic acid LC-ESI-QQ , negative-QTOF | splash10-03di-0000090000-b29692cfc9f66c4205c5 | 2017-09-14 | HMDB team, MONA | View Spectrum | | Experimental LC-MS/MS | LC-MS/MS Spectrum - Taurocholic acid LC-ESI-IT , negative-QTOF | splash10-0gvk-0019610000-6cd348047dc06e3abb7e | 2017-09-14 | HMDB team, MONA | View Spectrum | | Experimental LC-MS/MS | LC-MS/MS Spectrum - Taurocholic acid LC-ESI-QQ , positive-QTOF | splash10-014i-0010390000-b424218e255bf3091fda | 2017-09-14 | HMDB team, MONA | View Spectrum | | Experimental LC-MS/MS | LC-MS/MS Spectrum - Taurocholic acid LC-ESI-QQ , positive-QTOF | splash10-0a4i-0191200000-4fddfc0190213d6555ee | 2017-09-14 | HMDB team, MONA | View Spectrum | | Experimental LC-MS/MS | LC-MS/MS Spectrum - Taurocholic acid LC-ESI-QQ , positive-QTOF | splash10-08i9-0590000000-c210cb519c7e4192622e | 2017-09-14 | HMDB team, MONA | View Spectrum | | Experimental LC-MS/MS | LC-MS/MS Spectrum - Taurocholic acid LC-ESI-QQ , positive-QTOF | splash10-0w29-0900000000-42467048740170d1f693 | 2017-09-14 | HMDB team, MONA | View Spectrum | | Experimental LC-MS/MS | LC-MS/MS Spectrum - Taurocholic acid LC-ESI-IT , positive-QTOF | splash10-0002-0000940000-6c3a6cb6cef0d74f2139 | 2017-09-14 | HMDB team, MONA | View Spectrum | | Experimental LC-MS/MS | LC-MS/MS Spectrum - Taurocholic acid 10V, Negative-QTOF | splash10-03di-0000090000-21f22221a28d374a2518 | 2021-09-20 | HMDB team, MONA | View Spectrum | | Experimental LC-MS/MS | LC-MS/MS Spectrum - Taurocholic acid 10V, Positive-QTOF | splash10-03di-0000900000-bc321572fda51c26ea4b | 2021-09-20 | HMDB team, MONA | View Spectrum | | Experimental LC-MS/MS | LC-MS/MS Spectrum - Taurocholic acid 40V, Positive-QTOF | splash10-004i-2961000000-2ad87ae8f70b4deb4613 | 2021-09-20 | HMDB team, MONA | View Spectrum | | Experimental LC-MS/MS | LC-MS/MS Spectrum - Taurocholic acid 35V, Negative-QTOF | splash10-03di-0030090000-4366a02ee69b6b2b9a47 | 2021-09-20 | HMDB team, MONA | View Spectrum | | Experimental LC-MS/MS | LC-MS/MS Spectrum - Taurocholic acid 20V, Positive-QTOF | splash10-03di-0114900000-263e4adb5d4c8f0a9166 | 2021-09-20 | HMDB team, MONA | View Spectrum | | Experimental LC-MS/MS | LC-MS/MS Spectrum - Taurocholic acid 20V, Positive-QTOF | splash10-03di-0000900000-ffd27384b632f9e9db56 | 2021-09-20 | HMDB team, MONA | View Spectrum | | Experimental LC-MS/MS | LC-MS/MS Spectrum - Taurocholic acid 40V, Positive-QTOF | splash10-01p9-0389300000-86e8186c77b1ec43a6b7 | 2021-09-20 | HMDB team, MONA | View Spectrum | | Experimental LC-MS/MS | LC-MS/MS Spectrum - Taurocholic acid 10V, Positive-QTOF | splash10-014j-0000960000-cd066ae54dc0cc5a21fe | 2021-09-20 | HMDB team, MONA | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - Taurocholic acid 10V, Positive-QTOF | splash10-00ea-0601910000-36005a0ab81d9a5c22c7 | 2016-08-03 | Wishart Lab | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - Taurocholic acid 20V, Positive-QTOF | splash10-00di-2901200000-0277ee57e5a77bb559ed | 2016-08-03 | Wishart Lab | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - Taurocholic acid 40V, Positive-QTOF | splash10-006x-8903200000-6e18275cb262330c1d11 | 2016-08-03 | Wishart Lab | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - Taurocholic acid 10V, Negative-QTOF | splash10-03dj-3102890000-14b58b657187799f4abd | 2016-08-03 | Wishart Lab | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - Taurocholic acid 20V, Negative-QTOF | splash10-01qa-6404930000-b60c0ef5b779fc055945 | 2016-08-03 | Wishart Lab | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - Taurocholic acid 40V, Negative-QTOF | splash10-000x-9101100000-30f5c6ad01e5a5d282da | 2016-08-03 | Wishart Lab | View Spectrum |
NMR Spectra| Spectrum Type | Description | Deposition Date | Source | View |
|---|
| Predicted 1D NMR | 13C NMR Spectrum (1D, 100 MHz, H2O, predicted) | 2022-08-22 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 1H NMR Spectrum (1D, 100 MHz, H2O, predicted) | 2022-08-22 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 13C NMR Spectrum (1D, 1000 MHz, H2O, predicted) | 2022-08-22 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 1H NMR Spectrum (1D, 1000 MHz, H2O, predicted) | 2022-08-22 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 13C NMR Spectrum (1D, 200 MHz, H2O, predicted) | 2022-08-22 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 1H NMR Spectrum (1D, 200 MHz, H2O, predicted) | 2022-08-22 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 13C NMR Spectrum (1D, 300 MHz, H2O, predicted) | 2022-08-22 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 1H NMR Spectrum (1D, 300 MHz, H2O, predicted) | 2022-08-22 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 13C NMR Spectrum (1D, 400 MHz, H2O, predicted) | 2022-08-22 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 1H NMR Spectrum (1D, 400 MHz, H2O, predicted) | 2022-08-22 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 13C NMR Spectrum (1D, 500 MHz, H2O, predicted) | 2022-08-22 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 1H NMR Spectrum (1D, 500 MHz, H2O, predicted) | 2022-08-22 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 13C NMR Spectrum (1D, 600 MHz, H2O, predicted) | 2022-08-22 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 1H NMR Spectrum (1D, 600 MHz, H2O, predicted) | 2022-08-22 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 13C NMR Spectrum (1D, 700 MHz, H2O, predicted) | 2022-08-22 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 1H NMR Spectrum (1D, 700 MHz, H2O, predicted) | 2022-08-22 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 13C NMR Spectrum (1D, 800 MHz, H2O, predicted) | 2022-08-22 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 1H NMR Spectrum (1D, 800 MHz, H2O, predicted) | 2022-08-22 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 13C NMR Spectrum (1D, 900 MHz, H2O, predicted) | 2022-08-22 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 1H NMR Spectrum (1D, 900 MHz, H2O, predicted) | 2022-08-22 | Wishart Lab | View Spectrum | | Experimental 2D NMR | [1H, 13C]-HSQC NMR Spectrum (2D, 600 MHz, 100%_DMSO, experimental) | 2018-05-18 | Wishart Lab | View Spectrum |
|
|---|
| Biological Properties |
|---|
| Cellular Locations | |
|---|
| Biospecimen Locations | |
|---|
| Tissue Locations | - Fibroblasts
- Intestine
- Liver
- Neuron
- Placenta
|
|---|
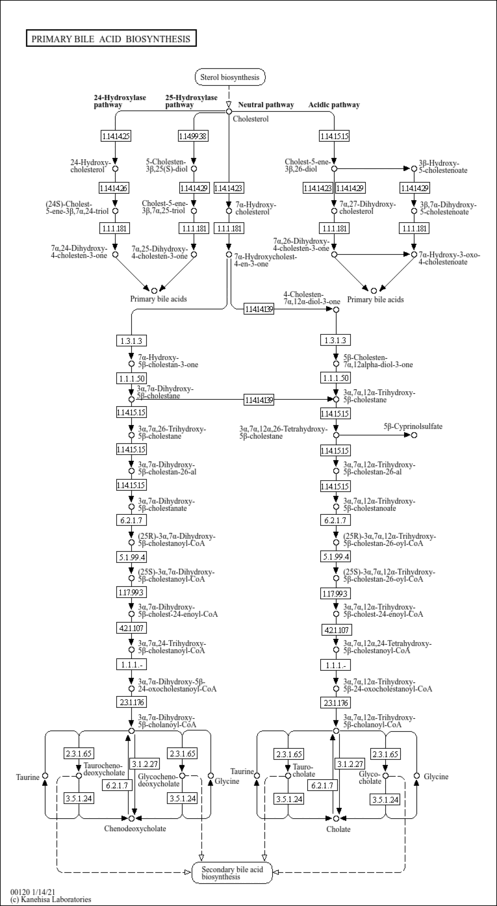
| Pathways | |
|---|
| Normal Concentrations |
|---|
| |
| Bile | Detected and Quantified | 9910 (9600-10220) uM | Adult (>18 years old) | Both | Normal | | details | | Blood | Detected and Quantified | 0.38 +/- 0.20 uM | Adult (>18 years old) | Both | Normal | | details | | Blood | Detected but not Quantified | Not Quantified | Adult (>18 years old) | Both | Normal | | details | | Blood | Detected and Quantified | 0.1(0.1-0.5) uM | Infant (0-1 year old) | Both | Normal | | details | | Feces | Detected and Quantified | 5.78 +/- 4.32 nmol/g dry feces | Not Specified | Not Specified | Normal | | details | | Feces | Detected but not Quantified | Not Quantified | Adult (>18 years old) | Both | Normal | | details | | Feces | Detected but not Quantified | Not Quantified | Children (1-13 years old) | Both | Normal | | details | | Feces | Detected but not Quantified | Not Quantified | Adult (>18 years old) | Both | Normal | | details | | Feces | Detected but not Quantified | Not Quantified | Adult (>18 years old) | Both | Normal | | details | | Feces | Detected but not Quantified | Not Quantified | Children (6 - 18 years old) | Not Specified | Normal | | details | | Feces | Detected but not Quantified | Not Quantified | Adult (>18 years old) | Both | Normal | | details | | Urine | Detected but not Quantified | Not Quantified | Adult (>18 years old) | Both | Normal | | details |
|
|---|
| Abnormal Concentrations |
|---|
| |
| Blood | Detected and Quantified | 9.8 +/- 1.9 uM | Infant (0-1 year old) | Both | GI disorder | | details | | Blood | Detected and Quantified | 5.6 +/- 1.6 uM | Infant (0-1 year old) | Both | GI disorder | | details | | Blood | Detected and Quantified | 1.2 +/- 0.5 uM | Infant (0-1 year old) | Both | GI disorder | | details | | Blood | Detected and Quantified | 0.2 +/- 0.1 uM | Infant (0-1 year old) | Both | GI disorder | | details | | Blood | Detected and Quantified | 0.8(0.3-2.7) uM | Infant (0-1 year old) | Both | Severe acute malnutrition | | details | | Blood | Detected but not Quantified | Not Quantified | Adult (>18 years old) | Both | Hepatocellular carcinoma | | details | | Blood | Detected but not Quantified | Not Quantified | Adult (>18 years old) | Both | Liver Cirrhosis | | details | | Feces | Detected but not Quantified | Not Quantified | Adult (>18 years old) | Both | CCD | | details | | Feces | Detected but not Quantified | Not Quantified | Adult (>18 years old) | Both | Ileal Crohn's disease | | details | | Feces | Detected but not Quantified | Not Quantified | Adult (>18 years old) | Both | Colorectal cancer | | details | | Feces | Detected but not Quantified | Not Quantified | Adult (>18 years old) | Both | Metastatic melanoma | | details | | Feces | Detected but not Quantified | Not Quantified | Not Specified | Not Specified | Recurrent Clostridium difficile infection | | details | | Feces | Detected but not Quantified | Not Quantified | Not Specified | Not Specified | Recurrent Clostridium difficile infection | | details | | Feces | Detected but not Quantified | Not Quantified | Adult (>18 years old) | Both | Clostridium difficile infection | | details | | Feces | Detected but not Quantified | Not Quantified | Adult (>18 years old) | Both | Colorectal Cancer | | details | | Feces | Detected but not Quantified | Not Quantified | Children (6 - 18 years old) | Not Specified | Crohns disease | | details | | Feces | Detected but not Quantified | Not Quantified | Children (6 - 18 years old) | Not Specified | Ulcerative colitis | | details | | Feces | Detected but not Quantified | Not Quantified | Children (6 - 18 years old) | Not Specified | Unclassified IBD | | details | | Urine | Detected and Quantified | 0.15 +/- 0.087 umol/mmol creatinine | Adult (>18 years old) | Both | Biliary atresia | | details |
|
|---|
| Associated Disorders and Diseases |
|---|
| Disease References | | Hepatocellular carcinoma |
|---|
- Ressom HW, Xiao JF, Tuli L, Varghese RS, Zhou B, Tsai TH, Ranjbar MR, Zhao Y, Wang J, Di Poto C, Cheema AK, Tadesse MG, Goldman R, Shetty K: Utilization of metabolomics to identify serum biomarkers for hepatocellular carcinoma in patients with liver cirrhosis. Anal Chim Acta. 2012 Sep 19;743:90-100. doi: 10.1016/j.aca.2012.07.013. Epub 2012 Jul 20. [PubMed:22882828 ]
| | Cirrhosis |
|---|
- Ressom HW, Xiao JF, Tuli L, Varghese RS, Zhou B, Tsai TH, Ranjbar MR, Zhao Y, Wang J, Di Poto C, Cheema AK, Tadesse MG, Goldman R, Shetty K: Utilization of metabolomics to identify serum biomarkers for hepatocellular carcinoma in patients with liver cirrhosis. Anal Chim Acta. 2012 Sep 19;743:90-100. doi: 10.1016/j.aca.2012.07.013. Epub 2012 Jul 20. [PubMed:22882828 ]
| | Colorectal cancer |
|---|
- Brown DG, Rao S, Weir TL, O'Malia J, Bazan M, Brown RJ, Ryan EP: Metabolomics and metabolic pathway networks from human colorectal cancers, adjacent mucosa, and stool. Cancer Metab. 2016 Jun 6;4:11. doi: 10.1186/s40170-016-0151-y. eCollection 2016. [PubMed:27275383 ]
- Goedert JJ, Sampson JN, Moore SC, Xiao Q, Xiong X, Hayes RB, Ahn J, Shi J, Sinha R: Fecal metabolomics: assay performance and association with colorectal cancer. Carcinogenesis. 2014 Sep;35(9):2089-96. doi: 10.1093/carcin/bgu131. Epub 2014 Jul 18. [PubMed:25037050 ]
| | Crohn's disease |
|---|
- Kolho KL, Pessia A, Jaakkola T, de Vos WM, Velagapudi V: Faecal and Serum Metabolomics in Paediatric Inflammatory Bowel Disease. J Crohns Colitis. 2017 Mar 1;11(3):321-334. doi: 10.1093/ecco-jcc/jjw158. [PubMed:27609529 ]
| | Ulcerative colitis |
|---|
- Kolho KL, Pessia A, Jaakkola T, de Vos WM, Velagapudi V: Faecal and Serum Metabolomics in Paediatric Inflammatory Bowel Disease. J Crohns Colitis. 2017 Mar 1;11(3):321-334. doi: 10.1093/ecco-jcc/jjw158. [PubMed:27609529 ]
| | Metastatic melanoma |
|---|
- Frankel AE, Coughlin LA, Kim J, Froehlich TW, Xie Y, Frenkel EP, Koh AY: Metagenomic Shotgun Sequencing and Unbiased Metabolomic Profiling Identify Specific Human Gut Microbiota and Metabolites Associated with Immune Checkpoint Therapy Efficacy in Melanoma Patients. Neoplasia. 2017 Oct;19(10):848-855. doi: 10.1016/j.neo.2017.08.004. Epub 2017 Sep 15. [PubMed:28923537 ]
| | Biliary atresia |
|---|
- Nittono H, Obinata K, Nakatsu N, Watanabe T, Niijima S, Sasaki H, Arisaka O, Kato H, Yabuta K, Miyano T: Sulfated and nonsulfated bile acids in urine of patients with biliary atresia: analysis of bile acids by high-performance liquid chromatography. J Pediatr Gastroenterol Nutr. 1986 Jan;5(1):23-9. [PubMed:3944741 ]
|
|
|---|
| Associated OMIM IDs | |
|---|
| External Links |
|---|
| DrugBank ID | DB04348 |
|---|
| Phenol Explorer Compound ID | Not Available |
|---|
| FooDB ID | FDB012335 |
|---|
| KNApSAcK ID | Not Available |
|---|
| Chemspider ID | 6423 |
|---|
| KEGG Compound ID | C05122 |
|---|
| BioCyc ID | Not Available |
|---|
| BiGG ID | 45150 |
|---|
| Wikipedia Link | Taurocholic_acid |
|---|
| METLIN ID | 5104 |
|---|
| PubChem Compound | 6675 |
|---|
| PDB ID | Not Available |
|---|
| ChEBI ID | 28865 |
|---|
| Food Biomarker Ontology | Not Available |
|---|
| VMH ID | TCHOLA |
|---|
| MarkerDB ID | Not Available |
|---|
| Good Scents ID | Not Available |
|---|
| References |
|---|
| Synthesis Reference | Schersten, Tore; Bjorntorp, Per; Ekdahi, Per H.; Bjorkerud, Soren. Synthesis of taurocholic and glycocholic acids by preparations of human liver. II. An analysis of the stimulating effect of the L fraction. Biochimica et Biophysica Acta, General Subjects (1967), 141(1), 155-63. |
|---|
| Material Safety Data Sheet (MSDS) | Download (PDF) |
|---|
| General References | - van Montfoort JE, Muller M, Groothuis GM, Meijer DK, Koepsell H, Meier PJ: Comparison of "type I" and "type II" organic cation transport by organic cation transporters and organic anion-transporting polypeptides. J Pharmacol Exp Ther. 2001 Jul;298(1):110-5. [PubMed:11408531 ]
- Rius M, Nies AT, Hummel-Eisenbeiss J, Jedlitschky G, Keppler D: Cotransport of reduced glutathione with bile salts by MRP4 (ABCC4) localized to the basolateral hepatocyte membrane. Hepatology. 2003 Aug;38(2):374-84. [PubMed:12883481 ]
- Hoekman MF, Rientjes JM, Twisk J, Planta RJ, Princen HM, Mager WH: Transcriptional regulation of the gene encoding cholesterol 7 alpha-hydroxylase in the rat. Gene. 1993 Aug 25;130(2):217-23. [PubMed:8359688 ]
- Sandker GW, Weert B, Olinga P, Wolters H, Slooff MJ, Meijer DK, Groothuis GM: Characterization of transport in isolated human hepatocytes. A study with the bile acid taurocholic acid, the uncharged ouabain and the organic cations vecuronium and rocuronium. Biochem Pharmacol. 1994 Jun 15;47(12):2193-200. [PubMed:7913319 ]
- Kullak-Ublick GA, Glasa J, Boker C, Oswald M, Grutzner U, Hagenbuch B, Stieger B, Meier PJ, Beuers U, Kramer W, Wess G, Paumgartner G: Chlorambucil-taurocholate is transported by bile acid carriers expressed in human hepatocellular carcinomas. Gastroenterology. 1997 Oct;113(4):1295-305. [PubMed:9322525 ]
- Claudel T, Inoue Y, Barbier O, Duran-Sandoval D, Kosykh V, Fruchart J, Fruchart JC, Gonzalez FJ, Staels B: Farnesoid X receptor agonists suppress hepatic apolipoprotein CIII expression. Gastroenterology. 2003 Aug;125(2):544-55. [PubMed:12891557 ]
- Rizzo G, Renga B, Mencarelli A, Pellicciari R, Fiorucci S: Role of FXR in regulating bile acid homeostasis and relevance for human diseases. Curr Drug Targets Immune Endocr Metabol Disord. 2005 Sep;5(3):289-303. [PubMed:16178789 ]
- Duan RD, Cheng Y, Tauschel HD, Nilsson A: Effects of ursodeoxycholate and other bile salts on levels of rat intestinal alkaline sphingomyelinase: a potential implication in tumorigenesis. Dig Dis Sci. 1998 Jan;43(1):26-32. [PubMed:9508530 ]
- Akao T: Influence of various bile acids on the metabolism of glycyrrhizin and glycyrrhetic acid by Ruminococcus sp. PO1-3 of human intestinal bacteria. Biol Pharm Bull. 1999 Aug;22(8):787-93. [PubMed:10480314 ]
- Jigorel E, Le Vee M, Boursier-Neyret C, Bertrand M, Fardel O: Functional expression of sinusoidal drug transporters in primary human and rat hepatocytes. Drug Metab Dispos. 2005 Oct;33(10):1418-22. Epub 2005 Jul 13. [PubMed:16014767 ]
- Zahner D, Eckhardt U, Petzinger E: Transport of taurocholate by mutants of negatively charged amino acids, cysteines, and threonines of the rat liver sodium-dependent taurocholate cotransporting polypeptide Ntcp. Eur J Biochem. 2003 Mar;270(6):1117-27. [PubMed:12631271 ]
- Wang LF, Luo H, Miyoshi M, Imoto T, Hiji Y, Sasaki T: Inhibitory effect of gymnemic acid on intestinal absorption of oleic acid in rats. Can J Physiol Pharmacol. 1998 Oct-Nov;76(10-11):1017-23. [PubMed:10100884 ]
- Yamamoto Y, Moore R, Hess HA, Guo GL, Gonzalez FJ, Korach KS, Maronpot RR, Negishi M: Estrogen receptor alpha mediates 17alpha-ethynylestradiol causing hepatotoxicity. J Biol Chem. 2006 Jun 16;281(24):16625-31. Epub 2006 Apr 10. [PubMed:16606610 ]
- Kwekkeboom J, Princen HM, van Voorthuizen EM, Meijer P, Kempen HJ: Comparison of taurocholate accumulation in cultured hepatocytes of pig, rat and man. Biochem Biophys Res Commun. 1989 Jul 31;162(2):619-25. [PubMed:2757635 ]
- Claudel T, Sturm E, Duez H, Torra IP, Sirvent A, Kosykh V, Fruchart JC, Dallongeville J, Hum DW, Kuipers F, Staels B: Bile acid-activated nuclear receptor FXR suppresses apolipoprotein A-I transcription via a negative FXR response element. J Clin Invest. 2002 Apr;109(7):961-71. [PubMed:11927623 ]
- Perwaiz S, Tuchweber B, Mignault D, Gilat T, Yousef IM: Determination of bile acids in biological fluids by liquid chromatography-electrospray tandem mass spectrometry. J Lipid Res. 2001 Jan;42(1):114-9. [PubMed:11160372 ]
- Johnson RC, Shah SN: Cholesterol ester hydrolase(s) in mammalian brain: is there a myelin-specific cholesterol ester hydrolase? Neurochem Res. 1986 Nov;11(11):1571-82. [PubMed:3683732 ]
- Balakrishnan A, Sussman DJ, Polli JE: Development of stably transfected monolayer overexpressing the human apical sodium-dependent bile acid transporter (hASBT). Pharm Res. 2005 Aug;22(8):1269-80. Epub 2005 Aug 3. [PubMed:16078136 ]
- Nittono H, Obinata K, Nakatsu N, Watanabe T, Niijima S, Sasaki H, Arisaka O, Kato H, Yabuta K, Miyano T: Sulfated and nonsulfated bile acids in urine of patients with biliary atresia: analysis of bile acids by high-performance liquid chromatography. J Pediatr Gastroenterol Nutr. 1986 Jan;5(1):23-9. [PubMed:3944741 ]
- St-Pierre MV, Kullak-Ublick GA, Hagenbuch B, Meier PJ: Transport of bile acids in hepatic and non-hepatic tissues. J Exp Biol. 2001 May;204(Pt 10):1673-86. [PubMed:11316487 ]
- Claudel T, Staels B, Kuipers F: The Farnesoid X receptor: a molecular link between bile acid and lipid and glucose metabolism. Arterioscler Thromb Vasc Biol. 2005 Oct;25(10):2020-30. Epub 2005 Jul 21. [PubMed:16037564 ]
- Chiang JY: Bile acid regulation of hepatic physiology: III. Bile acids and nuclear receptors. Am J Physiol Gastrointest Liver Physiol. 2003 Mar;284(3):G349-56. [PubMed:12576301 ]
- Davis RA, Miyake JH, Hui TY, Spann NJ: Regulation of cholesterol-7alpha-hydroxylase: BAREly missing a SHP. J Lipid Res. 2002 Apr;43(4):533-43. [PubMed:11907135 ]
- Williamson C, Gorelik J, Eaton BM, Lab M, de Swiet M, Korchev Y: The bile acid taurocholate impairs rat cardiomyocyte function: a proposed mechanism for intra-uterine fetal death in obstetric cholestasis. Clin Sci (Lond). 2001 Apr;100(4):363-9. [PubMed:11256973 ]
- Simons K, Toomre D: Lipid rafts and signal transduction. Nat Rev Mol Cell Biol. 2000 Oct;1(1):31-9. [PubMed:11413487 ]
- Watson AD: Thematic review series: systems biology approaches to metabolic and cardiovascular disorders. Lipidomics: a global approach to lipid analysis in biological systems. J Lipid Res. 2006 Oct;47(10):2101-11. Epub 2006 Aug 10. [PubMed:16902246 ]
- Sethi JK, Vidal-Puig AJ: Thematic review series: adipocyte biology. Adipose tissue function and plasticity orchestrate nutritional adaptation. J Lipid Res. 2007 Jun;48(6):1253-62. Epub 2007 Mar 20. [PubMed:17374880 ]
- Lingwood D, Simons K: Lipid rafts as a membrane-organizing principle. Science. 2010 Jan 1;327(5961):46-50. doi: 10.1126/science.1174621. [PubMed:20044567 ]
- Elshenawy S, Pinney SE, Stuart T, Doulias PT, Zura G, Parry S, Elovitz MA, Bennett MJ, Bansal A, Strauss JF 3rd, Ischiropoulos H, Simmons RA: The Metabolomic Signature of the Placenta in Spontaneous Preterm Birth. Int J Mol Sci. 2020 Feb 4;21(3). pii: ijms21031043. doi: 10.3390/ijms21031043. [PubMed:32033212 ]
- Gunstone, Frank D., John L. Harwood, and Albert J. Dijkstra (2007). The lipid handbook with CD-ROM. CRC Press.
|
|---|